January 2022: This page has been updated at Approximate Matching in the RxNorm API.
Changes in the algorithm (as of June 2015)
In the spring of 2015, the approximate matching algorithm as part of the approximatch match search function (REST: /approximateTerm) was revised. There were several reasons for doing this:- To decrease the time to process a request. In general, this function had a slow response time, with some requests taking a second or more to process. A heavy load of approxmimate match search requests could significantly slow down the system response time for all users.
- To correct some flaws in the algorithm that had been observed over time.
There are no changes to the input parameters or the output formats.
Below is a summary of changes that have been made to the algorithm, and some examples of the different results that will occur.
New normalization process. The normalization process was changed to replace NLM's Lexical Variant Generator (LVG)
norm function with the Lucene Porter Stemmer. This change greatly increases the speed of processing with minimal effect
on the results.
Spelling suggestion changes. Several changes were made to the spelling suggestion process. In the prior
version, only unknown strings that were at least six characters were spell corrected. In the new version, the minimum
length for spelling correction was reduced to five characters. Thus, "Aleev" will now be corrected to "Aleve". Another
significant change to the spelling corrections is the addition of multiple word corrections. For example, in the new
version the string "vitaminD" will be spell corrected with "vitamin D". The old algorithm did not permit multiple
word spelling suggestions.
Other changes. Several other minor algorithm changes were done to improve the speed. Some functional changes
such as adding additional abbreviations to the abbreviations table were done to improve the recognition of drugs.
Background
In September 2011, an approximate match string search function called approxMatch was added to the RxNorm API. This was the result of work done earlier as described in a paper presented at the 2011 AMIA Annual Symposium. In May 2013, a new function (REST: /approximateTerm) was added that provides additional output control and information. The following paragraphs describe the details of the approximate match functions in the RxNorm API.Purpose
The approximate match function finds the "closest" matches in the RxNorm data set with the input string. This function is useful for strings where an exact or normalized string match fails to return any results using /rxcui?name=.... For example, the following strings fail to be mapped to any concepts using/rxcui?name=...
:
ACCUPRIL 20 MG TAB TABLET (contains extra word) HYDROCHLOROT 50 MG TABLET (unknown abbreviation) Rantidine 15 ML Syrup Oral (misspelled word)Using the approximate match function will identify the top concepts that contain strings which most closely match the input string.
Details
Normalizing the input string
Each user string will be first normalized into tokens, using the RxNorm normalization approach
described in a AMIA paper.
The normalization process is linguistically motivated and involves stripping genitive marks,
transforming plural forms into singular, replacing punctuation (including dashes) with spaces,
removing stop words, lower-casing each word, breaking a string into its constituent words,
and sorting the words in alphabetic order. In addition, certain short forms are expanded, said forms are removed and numeric formatting is done.
Example:
Original term: METOPROLOL SUCCINATE 200MG TAB After RxNorm normalization: 200 metoprolol mg tablet
In the example, the RxNorm normalization expands tab into tablet,
separates 200 from mg, and removes the salt modifier succinate.
Then the string is lower cased and the words are sort alphabetically.
Identifying the drugs
After the user string is normalized, the approximate match algorithm
identifies the drugs in the user string. It compares each token with a list of drug names obtained from the
RxNorm ingredient and brand names. Once a drug is identified, all the strings in the data base containing that
drug become the candidate strings.
Example:
Original String: ACCUPRIL 20 MG TAB TABLET Drug identified: accupril Candidate strings from data base: Accupril Accupril Pill Accupril Oral Product Accupril 5 MG Oral Tablet Accupril 10 MG Oral Tablet Accupril 20 MG Oral Tablet Accupril 40 MG Oral Tablet quinapril 10 MG [Accupril] quinapril 5 MG Oral Tablet [Accupril] QUINAPRIL HYDROCHLORIDE 5 mg ORAL TABLET, FILM COATED [Accupril] (many more)In the example above,
ACCUPRIL
is identified as the drug by the approximate match function and all
strings containing
ACCUPRIL
are considered as candidates.If a token in the user string does not have a match in the data base, then the algorithm performs several tests to try and resolve the unknown token.
- Token splitting. If an unknown token contains both letters and numbers, the algorithm splits it into
tokens containing only letters and tokens containing only numbers.
EXAMPLE: unknown token Atripla600 is split into two tokens - Atripla and 600.
- Drug name expansion. The algorithm attempts to replace an unknown token with a drug name if the unknown token is a
shortened form of a drug.
EXAMPLE: unknown token HYDROCHLOROT is replaced with HYDROCHLOROTHIAZIDE.
NOTE: if the unknown token is a shortened form of more than one drug, drug name expansion is not done. - Spelling correction. If there exist unknown tokens after the above actions, then spelling correction
is attempted. The token must be at least five letters long. Only the top spelling suggestion (with ties)
based on edit distance is used.
EXAMPLE: unknown token Viagro will be replaced with the spelling suggestion Viagra.
In cases where no drug has been identified through the previous measures, a partial drug name match is attempted.
A candidate string list is created from tokens that are not associated with dosage or drug form words (such as
numbers, “mg”, “tablet”, “oral”, etc). This might occur if a multiple word drug name is underspecified.
Example:
User string: Penlac 8% oral solution No drug name found. After removing all dosage and drug form tokens, algorithm finds strings containing “Penlac”: Penlac Nail Lacquer Penlac Nail Lacquer 8% Topical Solution Penlac Nail Lacquer 80 MG/ML Topical Solution ciclopirox 80 MG/ML [Penlac Nail Lacquer] ciclopirox Topical Solution [Penlac Nail Lacquer] CICLOPIROX 80 MG TOPICAL SOLUTION [PENLAC] ciclopirox 80 MG/ML Topical Solution [Penlac Nail Lacquer] ciclopirox 80 MILLIGRAM In 1 MILLILITER TOPICAL SOLUTION [Penlac]
Scoring each candidate string
After the drugs have been identified, and the candidate strings containing the drugs have been extracted, the algorithm scores each string to determine the closeness to the user string. The tokens of each candidate string are compared to the tokens of the input variant string and the Jaccard’s coefficient is calculated to determine the similarity.
The score returned is a integer number between 1 and 100 inclusive which represents the Jaccard coefficient multiplied by 100 and rounded. The Jaccard coefficient is calculated by dividing the number of matching tokens in the input and candidate string over the union of the tokens of both strings.
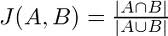
Example:
User string: Viagra 100 mg blue pill Candidate: Viagra 100 mg oral tablet # matched tokens: 3 (Viagra, 100, mg) # total tokens: 7 (Viagra, 100, mg, blue, pill, oral, tablet) Jaccard coefficient: 3/7 = 0.429 Score returned: 43
In May 2013, the scoring formula was modified to make spelling suggestions partial token matches.
The value of the partial
match will be either 0.75, 0.5 or 0.25 depending on how close the spelling suggestion is to the original token.
Example:
User string: abuticep Spelling correction: abatacept Partial match value: 0.25 Score returned: 25 User string: abuticept Spelling correction: abatacept Partial match value: 0.5 Score returned: 50 User string: abaticept Spelling correction: abatacept Partial match value: 0.75 Score returned: 75
Results returned from the API calls
The RxNorm API approximate match function /approximateTerm returns the score, rank, RxCUI and RxAUI of the closest strings. The string names are not returned due to the proprietary nature of some of the strings. The string names can be retrieved by calling /rxcui/{rxcui}/proprietary using the RxCUI and RxAUI as inputs to the function.
Also, /approximateTerm
returns a comment field which will indicate selected events such as spelling suggestions, token splitting,
drug name expansion and when no drugs are found. View comment messages
Examples
This section provides a number of examples illustrating the features of the algorithm discussed above. Note that in the output returned, the strings are added for clarity (only the score, rank, RxCUI and RxAUI are actually returned from the API call).input: chewable aspirin 81 mg tablet results: SCR R RXCUI RXAUI NAME 100 1 318272 3103140 ASPIRIN 81MG TAB,CHEWABLE 100 1 318272 1485034 Aspirin 81mg chewable tablet 100 1 318272 1485032 Aspirin Chew Tab 81 MG 100 1 318272 2639635 Aspirin 81mg Chewable tablet 100 1 318272 1485030 ASPIRIN 81MG TAB,CHEWABLE 100 1 318272 2836288 ASPIRIN 81MG CHEW TAB 100 1 318272 1485025 Aspirin 81 MG Chewable Tablet 100 1 318272 3517110 ASA 81 MG Chewable Tablet 100 1 318272 3103138 ASPIRIN 81MG CHEW TAB comment:In the above example, there are 9 strings with a top score of 100. Note that some of the strings contain abbreviations (CHEW, TAB) and acronyms (ASA) that are resolved by the algorithm.
input: chewable aspirn tablet 81 mg results: SCR R RXCUI RXAUI NAME 95 1 318272 3103140 ASPIRIN 81MG TAB,CHEWABLE 95 1 318272 1485034 Aspirin 81mg chewable tablet 95 1 318272 1485032 Aspirin Chew Tab 81 MG 95 1 318272 2639635 Aspirin 81mg Chewable tablet 95 1 318272 1485030 ASPIRIN 81MG TAB,CHEWABLE 95 1 318272 2836288 ASPIRIN 81MG CHEW TAB 95 1 318272 1485025 Aspirin 81 MG Chewable Tablet 95 1 318272 3517110 ASA 81 MG Chewable Tablet 95 1 318272 3103138 ASPIRIN 81MG CHEW TAB Comment: Spelling substitution: aspirin for aspirn;The input string above contains a spelling error which accounts for the lower top score than the previous example.
input: Bayer 81 mg results: SCR R RXCUI RXAUI NAME 60 1 794228 2802017 Aspirin 81 MG [Bayer Aspirin] 50 2 825181 2931865 Bayer Aspirin 81 MG Oral Tablet 50 2 825180 2931863 Bayer Aspirin 81 MG Chewable Tablet 43 4 825181 2969745 Bayer Low Dose, 81 mg oral tablet 43 4 825181 3857040 ASA 81 MG Oral Tablet [Bayer Aspirin] 43 4 825181 2931864 Aspirin 81 MG Oral Tablet [Bayer Aspirin] 43 4 825181 1167414 Bayer Low Strength, 81 mg oral tablet 43 4 794229 2802019 Bayer Aspirin 81 MG Enteric Coated Tablet 43 4 825180 3855698 ASA 81 MG Chewable Tablet [Bayer Aspirin] 43 4 825180 2931862 Aspirin 81 MG Chewable Tablet [Bayer Aspirin] Comment: Trying bayer as drug;In the above example, bayer is not a recognized drug (bayer aspirin is a brand name), but since no other drug was found, bayer is used as the drug and any database strings containing bayer become candidates.
input: HYDROCHLOROT 100 MG TABLET results: SCR R RXCUI RXAUI NAME 67 5 866479 1429164 Metoprolol & Hydrochlorothiazide Tab 100-25 MG 67 5 866479 2842481 HCTZ 25/METOPROLOL 100MG TAB 67 5 866491 2842512 HCTZ 50/METOPROLOL 100MG TAB 67 5 866491 3167842 HCTZ 50/METOPROLOL 100MG TAB 67 5 866479 3167811 HCTZ 25/METOPROLOL 100MG TAB 67 5 866491 1468220 Metoprolol & Hydrochlorothiazide Tab 100-50 MG Comment: Replaced hydrochlorot with hydrochlorothiazide;In the example above hydrochlorot is expanded to the ingredient hydrochlorothiazide. HCTZ is recognized as an acronym for hydrochlorothiazide.
input: tablet [EPC] results: SCR R RXCUI RXAUI NAME (none) comment: Trying epc as drug; Ambiguous top score (too many entries);In the above example, no drug is found, and epc is used to determine the drug candidates. This results in a large number of candidates with a top score, and the algorithm declares these results ambiguous and no data is returned.
input: XYZ oral tablet results: SCR R RXCUI RXAUI NAME (none) Comment: No drugs identified;The example above returns no results, and indicates in the comment returned that no drugs were identified. The token XYZ was not found in the database, otherwise a "Trying XYZ as drug" message would appear in the comment.


