Provides access to data collections created
to support research in consumer-health question answering,
extraction of adverse drug reactions, extraction of information
from MEDLINE®/PubMed® citations, and many other Lister Hill
National Center for Biomedical Communications, National Library
of Medicine (NLM) projects.
Go to the bioNLP resource
page
1,548 Consumer Health Questions submitted to NLM, de-identified and annotated with named entities from 15 broad categories, including medical problems, drug/supplements, anatomy, and procedures. For more information on this dataset, see Kilicoglu et al.(LREC 2016).
To help you standardize your units of measure, we’ve created this translation table that enumerates the UCUM syntax for many common unit patterns currently used in electronic reporting. We composed this early version in relatively short order and focused on the basics. It was based on content provided by Intermountain Healthcare, from a joint National Library of Medicine and Regenstrief Institute project analyzing raw units from more than 23 laboratory sources, and from the HL7 table of units. We excluded the units of measure for which we couldn’t find clear definitions or patterns of usage, those we believed would only be used in pharmacy dispensing, and units used for purely clinical reporting (e.g. cigarette pack-years). We have included most of the pure metric units from our sources, whether or not they apply directly to lab testing because they will be generally useful (and are pretty straightforward in UCUM).
- Dataset: http://loinc.org/usage/units
472 consumer health questions submitted to NLM, de-identified and annotated for spelling errors (non-word, real-word). For more information on this dataset, see Kilicoglu et al. (AMIA 2015).
URL: https://data.lhncbc.nlm.nih.gov/public/RIDEM/infobot_docs/CHQA_SpellCorrection_Dataset.zip
This file is an export of a key subset of the Panels and Forms represented in LOINC. The entire package of this key subset is currently available at http://loinc.org/downloads/accessory-files, in addition to separate packages for Laboratory panels, Clinical panels, Consumer Health panels, HEDIS panels, the HL7 Clinical Genetics panels, Newborn Screening panels, PhenX panels, US Government panels (including the CMS survey instruments MDSv2, MDSv3, OASIS, and CARE), and Other Survey Instruments. The hierarchical structure is represented in the file by the PARENT_ID, ID and SEQUENCE fields. The root, or top level, records in the file are those records where the PARENT_ID = ID. The records are in a Microsoft Excel spreadsheet (compressed as a zip file) with separate worksheets (tabs) for the form structure, LOINC code details, and answer lists.
Mapping your local laboratory test codes to LOINC can seem like a daunting task at first. Don't worry. To help you get started, we've created an empirically-based list of the most common LOINC result codes.
Knowing that relatively few codes account for much of the typical lab result volume, we think that this Top 2000+ list will be an excellent starter set. It contains just over 2,000 LOINC codes that represent about 98% of the test volume carried by three large organizations that mapped all of their lab tests to LOINC codes.
The LOINC Top 2000+ Lab Observations list is available in two varieties:
- US Version. For those who favor reporting in mass units (e.g. mg/dL)
- SI Version. For those who favor reporting in molar units (e.g. mmol/L)
To go with the Top 2000+ list, we've also written a Mapper's Guide that has a wealth of advice and guidance about which codes to choose for a given purpose. You can download it all here.
- Dataset: http://loinc.org/usage/obs
These 300 (or so) codes cover more than 95% of lab test orders in the U.S.
The LOINC Top 300 Lab Orders is a collection of universal laboratory order codes that covers the most frequent lab orders. It was created for use by developers of provider order entry systems that would deliver them in HL7 messages to laboratories where they could be understood and fulfilled. This value set was developed through both empirical and consensus-driven approaches. Obviously, at only 300 codes it doesn't include everything you might want to order, but is probably a very good "starter set". This is the Laboratory Order Value Set referenced by the HITSP C80 Clinical Document and Messaging Terminology Construct in (Table 2-96) and the current HL7 Version 2.5.1 Implementation Guide: S&I Framework Laboratory Orders from EHR, Release 1 being balloted in HL7 and developed in collaboration with the HHS S&I Framework Laboratory Orders Interface Working Group.
- Dataset: https://loinc.org/usage/orders/
Background:
Lung region detection in chest radiographs is an important early step in a machine learning (ML) pipeline for pulmonary disease screening and diagnosis. We are providing a dataset of lung masks with corresponding 55 frontal images (contrast enhanced) that have been subset from the NLM Open-i Indiana chest x-ray dataset (https://openi.nlm.nih.gov/faq#collection). This data set was first used in the publication below [1].
Reference:
- Xue Z, Yang F, Rajaraman S, Zamzmi G, Antani S, “Cross Dataset Analysis of Domain Shift in CXR Lung Region Detection”, Diagnostics 2023.
Includes the LOINC terms required to report all newborn screening results for all states — including variables for reporting an overall summary, for most of the card variables and, for reporting impressions, narrative guidance and measures of quantitative markers for each condition or condition category. Think of it as a master template from which each state can select the variables it needs to report NBS results in the same organizational structure.
- Dataset: https://loinc.org/54089-8
- Dataset in spreadsheet format (xls): http://newbornscreeningcodes.nlm.nih.gov/nb/sc/download/54089-8_Newborn_Screening_panel_AHIC-240.xls
- More guidance for e-reporting newborn screening results: http://newbornscreeningcodes.nlm.nih.gov/HL7
Includes the LOINC terms required to report all newborn screening results for all states — including variables for reporting an overall summary, for most of the card variables and, for reporting impressions, narrative guidance and measures of quantitative markers for each condition or condition category. Think of it as a master template from which each state can select the variables it needs to report NBS results in the same organizational structure. This same information in spreadsheet format can be imported into laboratory databases - http://newbornscreeningcodes.nlm.nih.gov/nb/sc/download/54089-8_Newborn_Screening_panel_AHIC-240.xls.
- Dataset: https://loinc.org/54089-8
- Learn More: https://lhncbc.nlm.nih.gov/newbornscreeningcodes/nb/sc/constructingNBSHL7messages.html
To help promote efficient electronic exchange of standard newborn screening data, the Lister Hill National Center for Biomedical Communications, in cooperation with the Newborn Screening Community and HITSP Population Perspective Technical Committee, developed draft guidance about the use of LOINC and SNOMED CT codes to report newborn screening test results in standard Health Level 7 (HL7) version 2.x message format.
- Annotated Example HL7 Message: https://lhncbc.nlm.nih.gov/newbornscreeningcodes/nb/sc/download/2014-09-02_NLM_HRSA_HL7_NBS_example_v6.pdf
- LOINC panel for Reporting Newborn Screening Results: https://loinc.org/54089-8
The Open-i project aims to provide next generation information retrieval services for biomedical articles from the full text collections such as PubMed Central. It is unique in its ability to index both the text and images in the articles. The article retrieval is powered by Essie (the search engine that supports ClinicalTrials.gov).
Open-i lets users retrieve not only the MEDLINE citation information, but also the outcome statements in the article and the most relevant figure from it. Further, it is possible to use the figure as a query component to find other relevant images or other visually similar images. Future stages aim to provide image region-of-interest (ROI) based querying. The initial number of images is projected to be around 600,000 and will scale to millions. The extensive image analysis and indexing and deep text analysis and indexing require distributed computing. At the request of the Board of Scientific Counselors, we intend to make the image computation services available as a NLM service.
Vist our Frequently Asked Questions page for more information and help.
Web Interface: https://openi.nlm.nih.gov/
Consumer Health Questions submitted to the Genetic and Rare Disease Information Center (GARD) manually labeled with question decomposition annotations.This includes sentence-level annotations (Question, Background, and Ignore), question-level annotations (Coordination, Exemplification), and a document-level annotation (Focus). For more information on this data, see Roberts et al. (LREC 2014; BioNLP 2014).
Dataset: Question Decomposition Data
Consumer Health Questions submitted to the Genetic and Rare Disease Information Center (GARD) manually labeled with question types. Uses the question decomposition annotations (above) to break multi-sentence questions into single-sentence sub-questions. Each sub-question has one question type designed to capture a high-level information need of a consumer health question (e.g., Diagnosis, Management, Susceptibility). For more information on this data, see Roberts et al. (BioTxtM 2014; AMIA 2014).
Downloads for LOINC Observations and Lab Orders Value Sets:
The following de-identified chest X-ray (CXR) image data sets are available to the research community along with findings and consensus radiologist annotations. Both sets contain normal as well as abnormal CXRs with the latter containing TB-consistent manifestations. The use and sharing of these deidentified images have been reviewed and exempted by the Ethics boards.
Please cite the following publications when using these data.
For datasets:
1. Jaeger S, Candemir S, Antani S, Wáng YX, Lu PX, Thoma G. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant Imaging Med Surg. 2014 Dec;4(6):475-7. DOI: 10.3978/j.issn.2223-4292.2014.11.20. PMID: 25525580; PMCID: PMC4256233.
For annotations:
2. Rajaraman S, Folio LR, Dimperio J, Alderson PO, Antani SK. Improved Semantic Segmentation of Tuberculosis-Consistent Findings in Chest X-rays Using Augmented Training of Modality-Specific U-Net Models with Weak Localizations. Diagnostics (Basel). 2021 Mar 30;11(4):616. DOI: 10.3390/diagnostics11040616. PMID: 33808240; PMCID: PMC8065621.
Montgomery County CXR Set: The images in this data set have been acquired from the TB Control Program of the Department of Health and Human Services of Montgomery County, MD, USA. This set contains 138 posterior-anterior CXRs of which 80 are normal and 58 are abnormal with manifestations that are consistent with TB. All images are de-identified and available along with left and right PA-view lung masks in PNG format. The data set also includes consensus annotations from two radiologists for 1024 × 1024 resized images and radiology readings. Download Montgomery County CXR Set
Shenzhen Hospital CXR Set: The CXR images in this data set have been collected and provided by Shenzhen No.3 Hospital in Shenzhen, Guangdong providence, China. The images are in PNG format. There are 326 normal and 336 abnormal CXRs, respectively, showing various TB-consistent manifestations. The data set also includes consensus annotations for a subset (N = 68) from two radiologists for 1024 × 1024 resized images and radiology readings. Download Shenzhen Hospital CXR Set
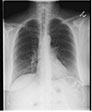
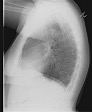
Background:
Chest radiographs are frequently taken as a screening and
diagnostic imaging tool. Very often, two views are taken. These
could be one of anterior-posterior (AP) or posterior-anterior
(PA) frontal views and a lateral view. Figure 1 shows examples
of frontal and lateral chest radiographs, respectively.
Appearance of anatomical structures is relatively unique in both
views. For computer-aided diagnosis (CAD) of cardiopulmonary
diseases several image processing and analysis steps are needed,
such as segmenting the lung region, locating relevant pathology,
and classifying it for diseases. For developing computer models
that can achieve these steps successfully and efficiently, there
is often a need to know the view of the chest X-ray. However,
the view information may be unavailable in the accompanying
metadata text or image headers due to poor image quality control
or aggressive deidentification.
(* The numbers of frontal and lateral images in the shared files (3864 and 3689 respectively) are different from those in [1], because not all images are publicly available.)
Reference:
- Xue Z, You D, Candemir S, Jaeger S, Antani SK, Long LR, Thoma GR, “Chest X-ray Image View Classification”, Proceedings of the 28th IEEE International Symposium on Computer-Based Medical Systems, 2015 (https://lhncbc.nlm.nih.gov/LHC-publications/pubs/ChestXrayImageViewClassification.html)
- D. Demner-Fushman, S.K. Antani, M. Simpson M, G.R. Thoma, “Design and development of a multimodal biomedical information retrieval system”, JCSE, vol. 6, no.2, pp.168-177, June 2012.
* https://openi.nlm.nih.gov/faq#collection
Download the Frontal view labels
Download the Lateral view labels