The MEDLINE N-gram Annaul Release Procedures
This page describes the procedures of generating annual release of the MEDLINE n-gram set:
N-Gram Directory
${MULTIWORDS_DIR}: ${LHC_GIT}/lexicon-multiwords
Pre-Process
- Source: the latest MEDLINE (released in middle Nov. for the next year) from MEDLINE/PubMed Baseline Repository
- Symbolic link ${MULTIWORDS_DIR}/data/Medline -> /nfsvol/nls/MEDLINE_Baseline_Repository (contact point: Jim Mork)
- copy xml files: ${MULTIWORDS_DIR}/data/Medline/${YEAR}/pubmedYYnXXXX.xml to ${DEV}/MedLine/data/${YEAR}/inData/PubMed.${YEAR}/.
- Symbolic link ${MULTIWORDS_DIR}/data/${YEAR}/outData/PubMed.${YEAR} -> ${DEV}/MedLine/data/${YEAR}/inData/PubMed.${YEAR}
Process
shell> cd ${MULTIWORDS_DIR}/bin
shell> 2.NGramGen ${YEAR}
| Steps | Description
|
|---|
| 1 (2017-) | Generate PmidTiAbSentences{YY}n{DDDD}.txt
Medline.GenPmidTiAbSentenceFiles.java
- Get all ${SRC}/medline${YY}n${NNNN}.txt to a list
- Sent to ${TAR}/PmidTiAbS${YY}n${NNNN}.txt by combine Ti and Tokenize Ab as sentence list
- 1 hr run time
|
| 1.1 (2018+) | Generate PmidTiAbS{YY}n{DDDD}.txt
Medline.GenPmidTiAbSentenceFileFromXmls.java
- Parse all ${MULTIWORDS_DIR}/data/${YEAR}/outData/PubMed.${YEAR}/pubmed${YY}n${NNNN}.xml to get PMID|TI|AB
- Use Java - StAX XML parser APIs
- StAX: Streaming API for XML
- not DOM (document object model) based, which allows random access to the document
- not SAX model: pushes data to application, with smaller memory and faster.
- Use streaming instead of DOM for smaller momory, use pull parsing insead of push parsing for better control.
- Combine and tokenize TI and AB of one citation (PMID) on the format of a list of sentences.
- Print to ${MULTIWORDS_DIR}/data/${YEAR}/outData/PmidTiAbSentence/PmidTiAbS${YY}n${NNNN}.txt
- ~400 files per hour
|
| Steps | Description
|
|---|
| 10 | Generate nGrams, with split option
|
| 11 | Group nGrams by alphabetic characters
|
| 12 | Filter by WC and combine alphabetic nGrams
|
| 13 | Sort nGrams
|
- Use GenAll script to generate n-gram automatically
shell> cd ${MULTIWORDS_DIR}/bin/2.NGramGenAll
- Edit files in ./inData/n-gram
- *** All above steps (10, 12, 13) are automatically done.
- *** Step: 11 requires manually setup.
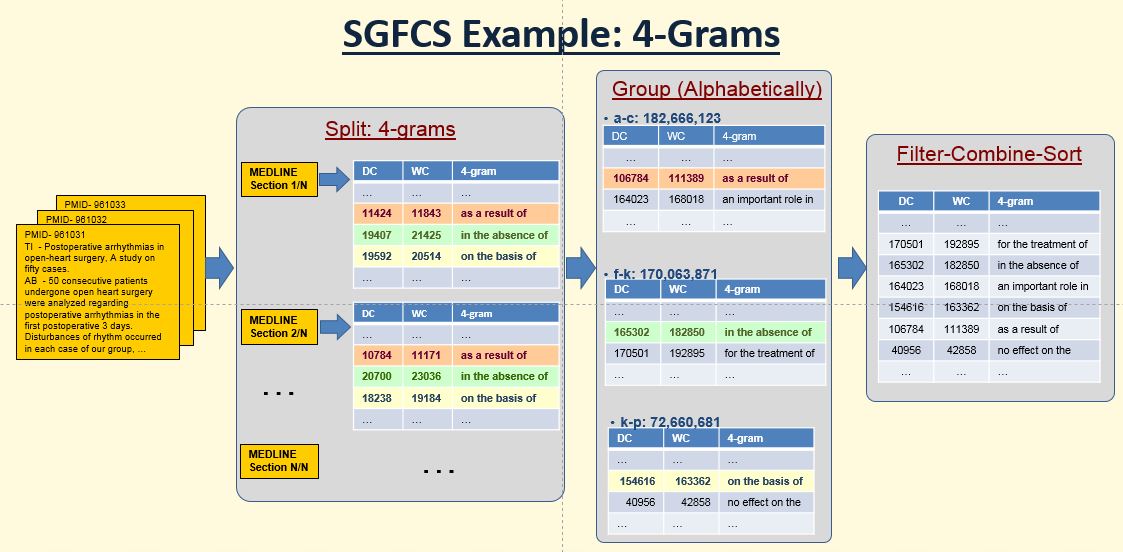
- Step-10 (Split):
the program automatically splits the MEDLINE once the nGram reaches certain number in HashMap (Must < 220,000,000). By adjusting this number, we can control the size (< 4.5 Gb) of each output split file and the performance time. Current set up is:
| 1-gram | 2-gram | 3-gram | 4-gram | 5-gram
|
|---|
| MaxHashMapSize | 150M | 150M | 150M | 130M | 120M
|
|---|
- Step-11 (Group):
*** requires manually setup the character alphabetical range (under ${MULTIWORDS}/bin/2.NGramGenAll/inData)
=> Use the following creteria for better performance:
- log files (${MULTIWORDS}/bin/02.NGramGenAll/logData/${YEAR}/${N}-gram/11-**.log from (this year and previous year) for a better guess
=> the nGram size (from log) should be around 150,000,000 (must < 220,000,000)
- This is the key factor for group's parameters.
- The Java HashTable run very slow when the key is close to 220,000,000
- The more gram, the bigger size of the file even the key is the same.
So, for 4-gram, 5-gram, use smaller key (nGram) size per file. The current setup is:
| 1-gram | 2-gram | 3-gram | 4-gram | 5-gram
|
|---|
| Max. Target HashMap Size (wc -l) | 50MB | 150MB | 170MB | 150MB | 130MB
|
|---|
| Max. Target File Size (ls -alh) | 1GB | 4GB | 5GB | 5GB | 5GB
|
|---|
=> It requires manually optimization to adjust the alphabetical range to fit into this range (very time comsuming).
=> Based on the detail log from previous year, try to arrange the range so that HashMap size and File size are within the target range.
run the setup similar to last year first, then split those with size greater than 5.0G by adjusting the neightber group.
those hifh frequency word, usch as of, the, on, in,me have high amount of n-grams.
=> Modification needs to be done in both:
- ${MULTIWORDS}/bin/2.NGramGenAll/inData/${YEAR}/${N}-gram/11-**.in
- ${MULTIWORDS}/bin/2.NGramGenAll/runGen${N}GramAll
- Size of output file in ${MULTIWORDS}/data/${YEAR}/outData/02.NGram/${N}-gram/2.Group:
=> All files from g01.NO-* ....g**.*-NO must be generated
=> n-gram size should be less than 220M (max. key size in Hash), it is better 50M ~ 150M.
=> file size should be around 4.0Gb (is better less than 4.5 - 5.5 Gb)
=> This value is just for reference.
=> the size depends on the ${N}.
- Log Files:
- ${MULTIWORD}/bin/Log.${YEAR}/02.NGramGen/
- log.sentence (log file for tokenizing PMID, TI, AB from Medline)
- log.1
- log.heap.n.50 (log file for generation n-grams)
- Move above log files to ${MULTIWORD}/bin/Logs/${YEAR}/02.NGramGen
- Add log.N to ${MULTIWORD}/bin/Logs/${YEAR}/02.NGramGen (for time spend)
- ${MULTIWORD}/bin/02.NGramGenAll/logData/${YEAR}
- n-gram/Step-option.log (log files for each options when using auto-program to generate n-grams)
shell> runGenNGramAll ${YEAR} > log.N
10
11=> takes time to manual configure for auto-run
12
13
===================
20
- need to set up options in the ./inData/${YEAR}
- log.N: for the running time on each step of each N-gram generation
- There are some Warning msg in step-10 regarding to not a good (sentence) tokenization, which is OK! These wraning msgs show on screen, not in the log.N file.
- Details (used to set the range in Step-11 for the next release):
- Post-Process
- Process Concept